|
|
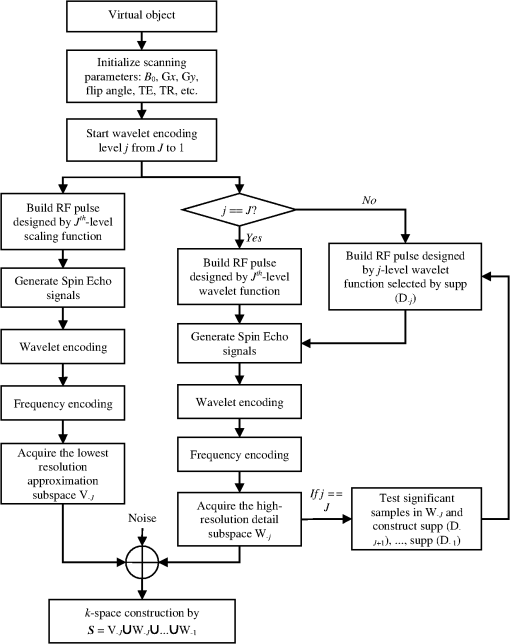
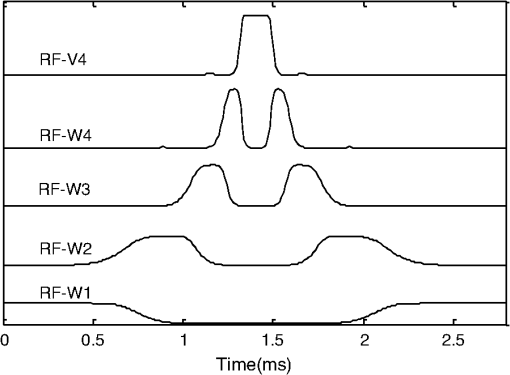
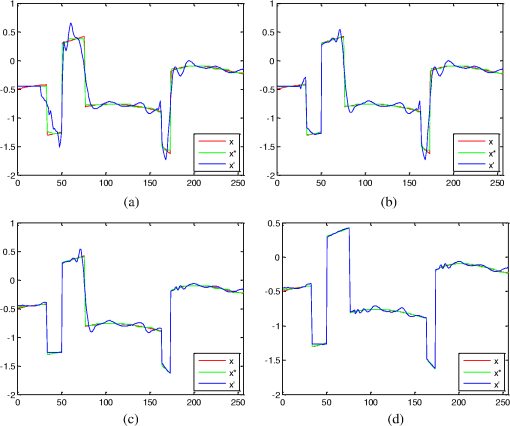
1.IntroductionIn conventional magnetic resonance imaging (MRI), scanners acquire samples of the encoded image in a spatial frequency (Fourier) domain, which is called -space. The MR image of the object in the spatial domain can be reconstructed through inverse Fourier transform of the -space data.1 A well-known problem in MRI is its long scan time that depends on the required number of acquired samples determined through Shannon-Nyquist sampling theory. Potential reconstruction of MR images from a reduced number of acquired samples without degrading the image quality would offer an effective method to reduce scan time while improving the resolution of current MR imagers. However, insufficient sampling of -space data violates the Shannon-Nyquist criterion and produces aliasing artifacts and noise in reconstructed MR images. Several common approaches to alleviate these artifacts and noise are to exploit redundancies in -space and to use temporal filtering, such as partial-Fourier, variable-density sampling, unaliasing by Fourier-encoding the overlaps using the temporal dimension (UNFOLD), etc.2–5 However, these methods still do not overcome low sampling rate artifacts. Investigations on the feasibility of wavelet encoding in MRI demonstrated better flexibility and adaptability in MR data acquisition than Fourier encoding.6–8 Unfortunately, the practicality of wavelet encoding in faster acquisition of MRI data has not been demonstrated in the above efforts. Through recent developments in the theory of compressive sensing (CS), MR images with a sparse representation in some transform domain, under the constraint of incoherent measurement, can be recovered from “randomly” under sampled -space.9,10 CS theory here exploits such sparsity of MRI data and reconstructs an MR image from very few incoherent measurements through a nonlinear procedure. Several methods have applied CS to Fourier-encoded MRI to reduce scan time.10–17 These methods are based on two characteristics of conventional MRI: MR images are naturally compressible in certain transform domains, and the sampling pattern is incoherent with respect to some sparsifying transformations. The natural fit of CS to MRI is discussed10 by reviewing the constraints required for successful reconstruction. Lustig et al.11 have applied CS to recover MR images from a subset of Fourier-encoded -space acquired by pseudo-random variable-density under sampling of phase-encoding to achieve fast imaging. The discrete cosine transforms (DCT), wavelet transform and finite-difference transform were used to exploit the sparsity of brain and angiogram MR images. The incoherence between these sparsifying transforms and Fourier operator were measured by the point spread function (PSF), and transform PSF (TPSF), whereas variable density random under sampling was used to improve the degree of incoherence. As their results showed, this CS approach achieved high reduction of imaging time in three-dimensional (3-D) Fourier-encoded imaging with good reconstruction performance. However, in two-dimensional (2-D) Fourier-encoded MRI, this approach does not achieve such benefit because only one-dimensioanl (1-D) sparsity was exploited. In mathematical CS theory, Fourier basis is maximally incoherent with the canonical basis.9,18 In traditional MRI, because the -space data are encoded by Fourier basis, the incoherent measurement constraint is only satisfied when the MR signal is sparse in the spatial domain. Consequently, it can be seen11 that CS-MRI is more suitable for angiogram imaging than for brain imaging because angiograms are sparse in their pixel representation. Therefore, Fourier-encoded MRI is not a good encoding scheme for stable and accurate CS reconstruction in most scenarios where MR signals are not sparse in the spatial domain. In addition, some existing crucial limitations remain unresolved in Fourier-encoded MRI, such as optimization of sampling trajectories and the computation time of reconstruction algorithms. Several studies have also investigated non-Fourier MRI and CS schemes to reduce the image acquisition time while improving the reconstruction quality.19–21 Random encoding of -space is achieved by tailoring spatially selective RF pulses to satisfy CS constraints in non-Fourier domains,19 achieving impressive reconstruction results. The CS constraints are satisfied better by making the energy of -space spread out in non-Fourier domain and pseudo 2-D random sampling.20,21 Despite the potential of the CS approaches in non-Fourier MRI to outperform traditional Fourier MRI, no general CS approach has yet been developed with strict theoretical justification. Recent research on application of CS to fast MRI suggests that there is no theoretical performance guarantee for CS-MRI with or without random encoding, and new CS based methods should be developed for specific applications.19 In recently published CS papers, adaptive sampling in the wavelet domain has been proposed to improve the signal recovery performance.22,23 Adaptive sampling schemes exploiting the tree structure of nonzero wavelet signal coefficients have been used to replace the ‘universal’ acquisition of random or pseudo-random sampling in traditional CS methodology.22,23 These schemes use nonrandom sampling and allow more control over the sensing procedure in the form of feedback to improve the CS performance significantly with fewer measurements and higher reconstruction quality. Haupt et al., have demonstrated that the adaptive sampling in CS, called compressive distilled sensing, has the advantage of significantly improving the error bounds compared to traditional CS schemes without adaptive sampling.24 These adaptive strategies have the potential to achieve more accurate and robust signal recovery in some practical applications.25 Therefore, if adaptive sampling is used in wavelet-encoded CS-MRI, it is possible to improve the reconstruction quality of images. We introduced well-known adaptive sampling in the wavelet domain, as used in image compression,26–28 to encode the MRI data yielding good reconstruction quality. In this paper, an efficient implementation of a specific CS approach in a simulator that allows good MRI reconstruction from a sparse wavelet-encoded -space following the well-known embedded zero-tree structure for image coding is presented.26–28 With this approach, sparsity of image data in the spatial domain is not required. In wavelet-encoded MRI, the -space data are encoded in multiple levels in the wavelet domain.6 The multilevel decomposed image is then transformed into a vector of sparse wavelet coefficients, and these few significant coefficients are encoded to reconstruct the image with high fidelity.26 Therefore, it is possible to represent the -space data with only a few significant samples and still reconstruct an MR image with a reduced scan time. Additionally, the reconstruction quality of such under sampled -space may be guaranteed by using the input vector of sparse wavelet coefficients in a specific CS framework. The following sections will discuss the proposed CS-MRI approach in the wavelet domain to achieve fast imaging and assess the reconstruction performance at different sampling rates, noise and sparsity levels. Section 2 introduces the procedure of sparse acquisition in wavelet-encoded MRI through the description of RF pulse definition, pulse sequence implementation and adaptive -space trajectories in a simulator. Section 3 presents an efficient CS algorithm based on the minimization of total-variation regularization and a least squares measurement of signal to reconstruct MR image from these sparsely encoded -space samples in the wavelet domain. Section 4 discusses the simulation results and reconstruction performance for phantom and brain images. Section 5 describes feasibility of future applications of our work. 2.Implementation of Sparse Data Acquisition in Wavelet-Encoded k-Space2.1.Sparsity of Image/Signal in Wavelet DomainThe wavelet transform of natural images produces a large number of coefficients of values with zero or near-zero magnitudes and a small number of significant values (e.g., the ones with larger magnitude than a determined threshold ). If we localize and encode these significant values, it is possible to reconstruct images with high fidelity. In addition, wavelet transform provides a compact multiresolution representation of the image, and the significant wavelet coefficients are well-organized in hierarchical trees. Thus, the high-resolution detail tends to be significant only if significant details can be found at all resolutions from the lowest level. This property is also commonly used to predict the positions of significant information across scales in wavelet-based compression algorithms.26–28 Therefore, for many natural or medical images, it is possible to represent the wavelet-encoded -space using only a few tree structured significant samples. These samples capture most of the signal information and can be used to reconstruct the MR image with insignificant distortion. This sparse acquisition of wavelet-encoded -space data is achieved by designing spatially selective RF excitation pulses for generating adaptive -space trajectories with reduced scan time. 2.2.Wavelet-Encoded MRIAcquisition of MRI data in the wavelet domain has been investigated by a number of researchers7,8 to provide more flexibility and adaptivity than traditional Fourier-encoded MRI. Wavelet-encoding is used to replace phase-encoding, and the wavelet-encoded -space is represented in multiple levels. The low-resolution approximation subspace is spanned by a family of scaling functions, and the high-resolution detail subspace is spanned by a family of wavelet functions. The -space can be reconstructed through these subspaces, In particular, assume that -axis is the wavelet encoding direction and -axis is the frequency encoding direction. In each subspace, the resolution of the -axis is constant, and the resolution of the -axis is varied with the decomposition level . In the -level subspace, the resolution along the -axis can be calculated by , where is the highest resolution when . In order to simplify the discussion, we use to represent one line of fully sampled frequency encoding at the location , e.g., . Therefore, the wavelet-encoded MRI can be considered as wavelet transform of one dimension signal . For level , each subspace is constructed by wavelet coefficients as follows: and Here, , and , . The coefficient set in in terms of the scaling function represents a coarse approximation of the signal. The coefficient sets {, } in terms of the wavelet function {, } represent the detail or difference between the coarse approximation and a finer resolution approximation. From Eqs. (2) and (3), we observe that the sparse acquisition of a signal can be achieved by tailoring spatially selective RF excitation pulses.A hierarchical binary tree structure exists in the wavelet coefficients set, where all wavelet coefficients can be arranged by a “parent–child” relationship.28,29 The coefficient in is called “parent,” and all coefficients corresponding to the same spatial location in {, } are called “child.” If the “parent” is insignificant, all its “children” are insignificant. All “parents” are tested first for significance, and then the positions of all significant “children” are contained in their trees. It is proposed that the wavelet-encoded MRI data be acquired starting with the lowest resolution -space and then the next finer scale -space {, } be undersampled based on the tested results in to acquire only significant samples. The support of coefficient set is defined to be the positions where its coefficients are larger in magnitude than an application dependent threshold . The support of is thus represented by Here, determines the number of samples in -space. According to the support of , we could calculate the support of coefficients {, } corresponding to the tree structure, denoted as {, }.2.3.Simulation of Sparse MR Data Acquisition in the Wavelet DomainMost MRI simulators are developed based on the solutions of the Bloch equation, and the generated MR signal is encoded in Fourier domain.30,31 Wavelet domain simulators were developed in earlier years but did not demonstrate any advantage in MRI scanning time.6–8 In this paper, a simulator generating MR signal in wavelet domain using adaptive sampling in a CS framework has been developed by designing RF excitation pulses and pulse sequences as described in detail in the following sections. This simulator generating MR signals in the wavelet domain is developed for sparse data acquisition as shown in Fig. 1. The executable code in MATLAB for our simulator can be accessed from the following link: http://www2.ece.ttu.edu/CVIAL/simulators/WaveletCSMRISim.exe. Our simulator is a modified version of a Fourier domain MRI simulator developed by Yoder et al. in 2004.30 2.3.1.Simulator overviewAn overview of the proposed MRI simulator to acquire sparse wavelet-encoded -space data is shown in Fig. 1. In the proposed data acquisition scheme, we do not need the information from fully sampled data. We actually acquire the wavelet-encoded MRI data starting with the lowest resolution -space by designing spatially localized wavelet shaped RF excitation pulses for generating -space trajectory by replacing the traditional phase encoding trajectory. Only the lowest resolution trajectory is fully sampled (requiring a small number of RF excitation pulses and the least data acquisition time) to identify for the significant parent wavelet coefficients and to allow full sampling across the levels if desired. The subsequent finer scale -space {, } data are generated by a multiple (e.g., two to eight for four level wavelet encoding based on the parent–child tree structure of wavelet coefficients) of RF excitation pulses as shown in the encoding overview in Fig. 1 (see Sec. 4.2 for details of implementation). Using a virtual object, scanning parameters, such as , , (magnetic gradient along and axis), flip angle, TE (time echo), TR (time repetition), are initialized first. This simulator generates -space data in multiple levels. For -level imaging, the level of -space acquisition starts from to 1. When equals to , two RF excitation pulses, which are designed, respectively by the ’th-level scaling and wavelet functions, are used to generate spin echo signals. After wavelet-encoding and frequency-encoding, the subspaces and are acquired, and then significant samples are tested in to construct {, }. When level is from to 1, RF excitation pulses are designed by ’th-level wavelet function and indexed by supp () to acquire under sampled -space W-j. Finally, the sparse-encoded -space is constructed by these subspaces , and to to contain all significant samples. To simulate realistic images, noise is added to the -space. 2.3.2.Design of RF excitation pulsesExcitation profiles in this wavelet-encoded MRI simulator are varied and shaped by functions of wavelet transform. At small flip angles (less than 30 deg), RF pulses are shaped by Fourier transform of excitation profiles. Here, we utilize the Battle-Lemarie wavelet basis functions as shown in Fig. 2 to design RF pulses. Figure 2(a) shows the wavelet scaling and basic functions and in the spatial domain. The major reason to choose this specific basis function set is that their Fourier transforms are smooth and rapidly decay to zero as shown in Fig. 2(b). This basis set provides short RF pulses with relatively precise spatial excitation profiles.6 Fig. 2Battle-Lemarie functions. (a) Scaling and wavelet basis functions in time domain; (b) Fourier transforms of scaling and wavelet functions. 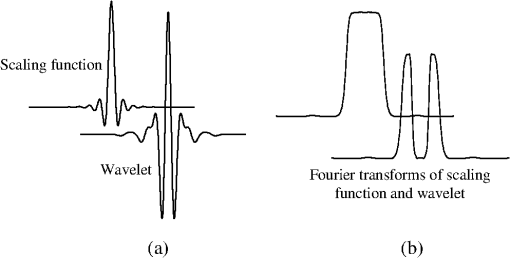 For -level wavelet encoding, assume that the gradient strength along -axis is , the FOV along -axis is centimeters, and the highest resolution is . According to Eqs. (1)-(3), we combine one set of scaling functions {, } and sets of wavelets {, ; } required to shape the excitation profiles. Therefore, () RF pulse profiles and {, } are required to generate -spaces and {, }, where and {, } are Fourier transforms of and {, }, respectively. The excited location along -axis is determined by the center carrier frequency of the RF pulse. In order to cover the entire FOV, the fundamental size of translation step is . According to the Bloch equation, there is a linear mapping of the resonant frequency of the spins and the spatial location , i.e., , and the fundamental frequency step , where is gyromagnetic ratio (approximately for hydrogen). For small-flip-angle excitation, the carrier frequencies of each pulse in {} and {} are offset by {} and {} correspondingly to excite each of the locations {} and {}. The duration time of pulse {} is computed by and used to compute the half-power width of {}. For example, in 4-level wavelet encoding (e.g., ), , , and . Five types of pulse profiles, labeled RF-V4, RF-W4, RF-W3, RF-W2 and RF-W1, are required to produce these RF pulses and {, }. The fundamental frequency step is 426 Hz, and the duration time of pulse RF-V4 is approximately 0.15 ms, as shown in Fig. 3. After reconstruction of MR images in the spatial domain, the imaging resolution of 1 mm can be achieved. Any deviation of (e.g., inhomogeneity along -axis) would result in a distortion of excitation profiles, especially occurring in high static field () and with tailored RF pulses. In practice, this distortion can be corrected using a prescan with the same sequence. A fully sampled image acquired for each spatially selective pulse is used as a standard to compute compensation parameters for each excitation profiles.19 The inhomogeneity maps generated from 3-D images are acquired by prescans and the carrier frequency and amplitude of pulses are adjusted to excite a uniform slice.32 In this paper, if the inhomogeneity maps (e.g., ) is acquired using prescans, the distortion of excitation profiles can be calibrated by adjusting the carrier frequency of each pulses. In the simplified version of this simulator, we neglect the inhomogeneity in and there is no distortion of excitation profiles. 2.3.3.Pulse sequencesDuring MR imaging, the timing of pulse sequences determines the RF signal acquisition and -space trajectories. Pulse sequences contain RF pulses and magnetic field gradients. In wavelet-encoded MRI simulator, the timing of pulse sequences is defined with spatially selective RF excitation and adaptive -space trajectories to yield a sparse encoding scheme. As shown in Fig. 1, at the beginning, RF pulses {, } and {, } with the carrier frequency offset by {} are used to generate -level wavelet-shaped excitation profiles along axis. In the following step, a precession with application of magnetic gradient along axis is specified by its duration and the gradient magnitude to fill the ’th line of frequency-encoding in -space. After RF pulses and lines of frequency-encoding, the -space and are fully sampled. In , the positions of significant samples, , are tested by Eq. (4) and then {, } are constructed. When is from to 1, RF pulses {, } with the carrier frequency offset by {} and magnetic gradient are applied to achieve the trajectory consisting only of significant samples in . Finally, the sparsely encoded -space constructed by these subspaces , to contains the significant samples. 2.3.4.NoiseIn real MRI scanners, the acquired raw -space data are considered to be corrupted by complex white Gaussian noise with the same variance in the real and imaginary parts. After reconstruction of MR images from the corrupted -space data, the noise embedded in the image intensity has Rayleigh statistics in the background and Rician statistics in the signal regions.33,34 Therefore, this noise model is used in this simulator with different signal to noise ratio (SNR), with SNR is defined as where is the average root mean square (RMS) amplitude of the signal, and is the average RMS amplitude of the noise.3.CS Reconstruction of MR images3.1.CS Scheme in Wavelet-Encoded MRIIn the proposed wavelet-encoded MRI simulator, if -space data are fully sampled based on Shannon-Nyquist sampling theory, an MR image could be reconstructed by inverse wavelet transform along the wavelet-encoding direction and inverse Fourier transform along the frequency-encoding direction. If -space data are undersampled, this method cannot reconstruct MR images accurately. CS theory offers a potential to reconstruct a compressible signal from far fewer samples than that required by Shannon-Nyquist sampling theory and provides a new approach to overcome conventional limitations in signal sampling.9,18 Therefore, CS may be applied to reconstruct MR image from under sampled -space. The reconstruction quality of such under sampled -space is guaranteed by using the input vector of sparse wavelet coefficients in a specific CS framework. Because the frequency encoding is fully sampled, if is recovered, MR image can be reconstructed by inverse Fourier transform of . The main task of CS reconstruction is to reconstruct from the sparse wavelet-encoded -space replacing the traditional phase encoding direction. According to Eq. (2) and (3), and can be represented as and where is the matrix whose rows are for , and is the matrix whose rows are for . We denote ,, and . Therefore, the measurement of wavelet-encoded -space can be represented as where is the sensing matrix, is the identity matrix, is a rectangular matrix that selects elements of any ()-dimensional vector () and is determined by {supp , }. We consider the global sensing matrix and the sparse wavelet coefficients set in Eq. (8) to be Because the sensing matrix involves partial wavelet transform, it is not suitable to exploit the sparsity of by wavelet coefficient set due to the “incoherent” constraint in the CS framework. Most real MR signals are approximately piecewise smooth and have a small total variation (),11,19 which is defined as . The use of regularization could promote sparsity of finite differences in MR images and reduce ringing artifacts near edges that are caused by tailoring of the wavelet coefficients.8 Therefore, the reconstruction of , denoted as , can be computed by solving the following problem:35 where -norm is defined as and parameter controls the fidelity of to the measurements and is determined by estimating the noise variance. Nesterov’s algorithm can be used to solve the Lagrangian form of (10) to achieve the reconstruction by 36 where is used to balance regularization, and the measurement consistency is experimentally set. Large leads to staircase effect. To solve the CS problems in Eq. (11), the CVX software package by Grant, Boyd, and Ye was used ( http://www.stanford.edu/~boyd/cvx). Although the CVX package is a slow code, we can acquire the raw -space data first and then implement reconstruction off-line. This procedure still reduces the scanning time and provides the potential to achieve fast imaging.3.2.Reconstruction StabilityIn order to guarantee the performance of reconstruction by Eq. (10), the global sensing matrix in Eq. (9) must satisfy the restricted isometry property (RIP).37,38 If satisfies the RIP of order , for all -sparse vectors , where is the isometry constant, which should be a small number to satisfy RIP constraint. Because the degree of coherence may be considered as isometry constant of order 2, the RIP-based guarantees are generally stronger than incoherence-based guarantees for stable and accurate reconstruction.39 The reconstruction performance is improved as is made smaller. For example, from recent CS papers,38–40 if in a noiseless environment, the -sparse vector can be reconstructed exactly by Eq. (11) with with fewer than nonzero entries. However, given any matrix , it is often infeasible to compute practically useful RIP-based guarantees for all sparse vectors due to computational problems. Therefore, we investigate the RIP guarantees for , which is designed adaptively based on a sparse vector .In Eq. (8), the measurement is divided into two parts and , where is the fully sampled scaling coefficient set and is the under sampled wavelet coefficient set . Therefore, the measurement consistence constraint equals to , and the RIP condition in Eq. (12) can be rewritten as Larger values of result in smaller . For example, if , . Thus, all nonzero entries in are contained in the under sampled -space , and can be recovered exactly. Therefore, sparse acquisition of wavelet-encoded -space data can result in a smaller value of than other encoding methods by including as many significant samples as possible in such under sampled -space. In addition, for each MR image, specific global sensing matrix is constructed adaptively by investigation of sparsity in wavelet-encoded -space. 3.3.CS Reconstruction ExperimentThe numerical experiments for CS reconstruction of 1D piecewise smooth signal {, } are performed by 4-level sparse wavelet encoding. The proposed CS reconstruction result from significant wavelet coefficients only, denoted as , is compared with the best sparse approximation result, denoted as , which includes the zero-filled insignificant coefficients as well. Figure 4 shows the reconstruction results and with different measurement number , 88, 116 and 144. The measurement at each sampling number is acquired by sparse encoding in the wavelet domain. CS reconstruction performs better than the best sparse approximation reconstruction does with approaching the original signal closely under all conditions. Table 1 shows RIP constants and reconstruction SNRs at different measurement number . Larger values of result in smaller values of and higher reconstruction SNRs by CS with better sparse approximation results. Table 1The RIP constants and SNRs for each sampling number.
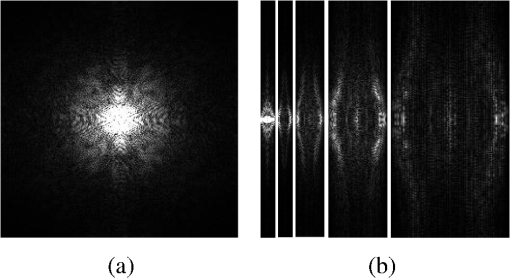
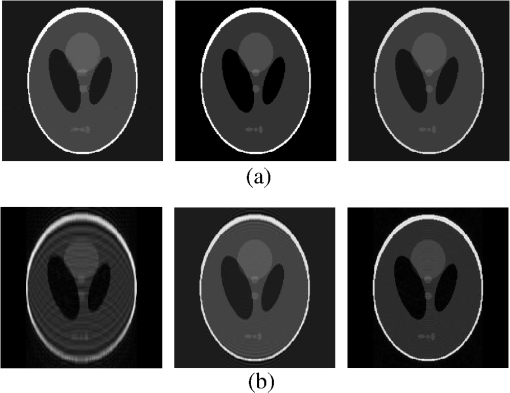
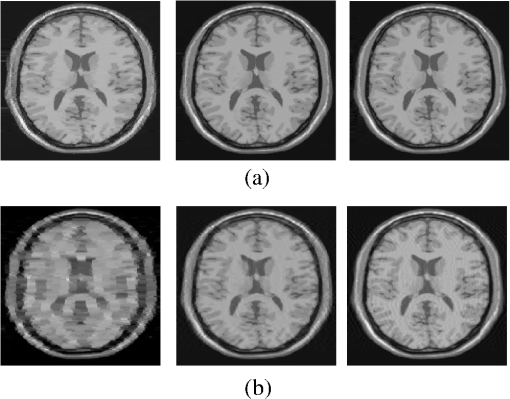
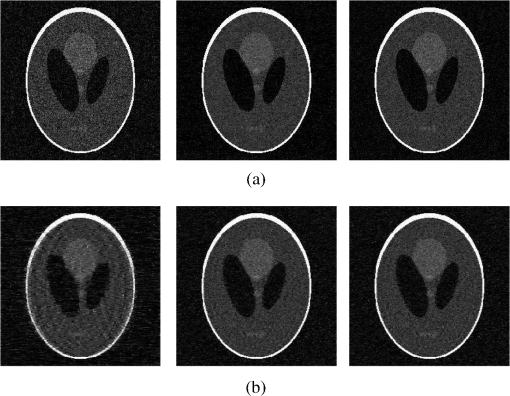
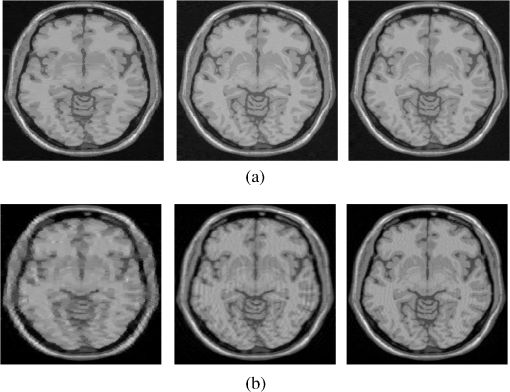
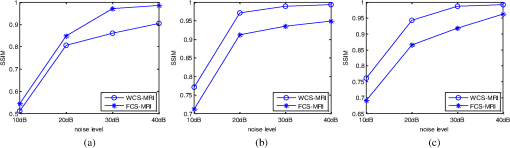
4.Experimental ResultsThe Shepp-Logan phantom and a 3-D digital brain phantom from the McConnell Brain Imaging Center, Montreal Neurological Institute, McGill University are used as virtual objects in the MRI simulator to investigate the properties of the proposed wavelet-encoded CS-MRI scheme (WCS-MRI).41,42 The digital brain phantom has a spatial resolution of and contains most of the relevant tissue types. Based on these phantoms, we first simulate the fully sampled wavelet-encoded MRI, and then we simulate the sparse acquisition of -space data at different sampling rates and reconstruct MR images by CS. These simulation results are compared with the results of the commonly used Fourier-encoded CS-MRI scheme (FCS-MRI). In Fourier-encoded -space, most of the energy is concentrated close to the center and rapidly decays toward the periphery. Therefore, a variable-density sampling scheme is implemented in FCS-MRI, where the phase encoding locations are randomly spaced but cover the low-frequency portion near the center of -pace, matching the energy distribution in -space. The CS reconstruction of MR image is performed from the undersampled -space data. 4.1.Implementation of Wavelet-Encoded MRI SimulationData are collected for MR image reconstruction on a voxel grid using the Shepp-Logan phantom and two different brain slices (slice 1 and slice 2) from the brain phantom. Figures 5 and 6 show fully sampled Fourier-encoded and 4-level wavelet-encoded MRI results, respectively. These results are generated with zero noise and homogeneous magnetic field (). The imaging parameters are , and . For each image, there are 256 RF excitation pulses required, and its spatial resolution is . It is seen visually that wavelet-encoded MRI acquires images with the same quality and resolution as Fourier-encoded MRI does. The quantitative assessment of the quality of the reconstructed images has been computed by peak signal to noise ratio (PSNR) and structural similarity (SSIM) as shown in Sec. 4.2. Fig. 5Fully sampled Fourier-encoded MR images of (a) the Shepp-Logan phantom, (b) brain image 1 and (c) brain image 2. 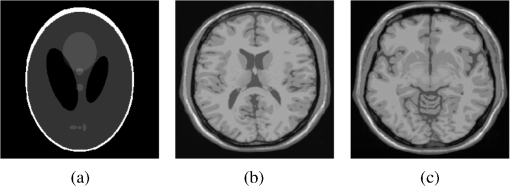 Fig. 6Fully sampled 4-level wavelet-encoded MR images of (a) the Shepp-Logan phantom, (b) brain image 1 and (c) brain image 2. 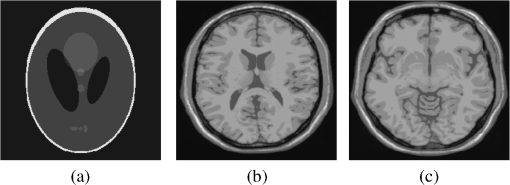 Figure 7 shows the energy of -spaces acquired by Fourier- and wavelet-encoded MRI for brain image 1, respectively. In Fig. 7(a), most of the energy in Fourier-encoded -space is concentrated close to the center of -space, at low spatial frequencies. However, in Fig. 7(b), the wavelet-encoded -space is represented in multiple levels with a high degree predictability between levels. The coarse scale subspace has its spectrum localized in the low frequency band, while these fine scale subspaces to have their energy widely dispersed in the high frequency band and well-organized in trees along wavelet encoding direction. 4.2.Simulation of CS-MRIThe proposed WCS-MRI scheme has been simulated with the phantoms shown in Fig. 5 with different sampling rates. The implementations of sampling patterns reported22–24 are not the same as our approach. Our data acquisition time is reduced by sparse acquisition of only significant samples across multiple levels by designing appropriate RF excitation pulses at each level. Such wavelet-encoded MRI provides the flexibility and adaptivity to represent the -space data, especially in multiple levels. According to the wavelet decomposition theory, it is possible to achieve the sparse acquisition of -space data by exploiting the hierarchical binary tree structure in the wavelet coefficients set.6–8,28 Our implementation is based on the structural characteristics of wavelet coefficients and uses a desired percentage of the full samples in our proposed CS-MRI making the acquisition time directly proportional to the sampling rate. Our data acquisition time is reduced by sparse acquisition of only significant samples across multiple levels by designing appropriate RF excitation pulses at each level. For 4-level wavelet-encoded MRI, there are 16 RF excitation pulses required to generate and , respectively. If significant samples in are generated by only RF pulses (), the corresponding number of RF pulses in , and are , and , respectively. Therefore, the total sampling rate (SR) can be calculated by , including 32 samples from and . In this simulation, if , the corresponding sampling rate . The scan time is determined by SR, where is the imaging time in fully encoded MRI with the same resolution. Finally, MR images are reconstructed from these under sampled -spaces through CS. In order to compare with WCS-MRI, the same number of RF pulses is used to acquire -space data in FCS-MRI. To make the simulation close to real MRI acquisition, Gaussian noise is added. 4.2.1.Simulations under ideal conditionsFigures 8 and 9 show the reconstruction of test images by WCS- and FCS-MRI schemes without noise; sampling rates are 23%, 40%, and 56% (e.g., 60, 102 and 144 RF excitation pulses used), respectively. Large-scale ringing artifacts are apparent at low sampling rate (e.g., ) in the FCS-MRI results. However, artifacts in WCS-MRI are much less than those in FCS-MRI, and more detail and high frequency components are kept in WCS-MRI than in FCS-MRI. When the sampling rate is 40%, no artifacts are apparent in WCS-MRI, while artifacts are still visible in FCS-MRI. When the sampling rate is 56%, both reconstruction schemes perform well without apparent artifacts. Because the Shepp-Logan phantom image is much sparser in pixels than brain images, CS reconstruction of the Shepp-Logan phantom image can achieve a much better performance. 4.2.2.Simulations under noisy conditionsSimulations are also performed to illustrate reconstruction performance with noise. Figures 10 and 11 show low-SNR simulation results (e.g., ) of the Shepp-Logan phantom reconstruction and high-SNR simulation results (e.g., ) of brain image 2 reconstruction by WCS- and FCS-MRI schemes with sampling rates 23%, 40%, and 56%. In Fig. 10, FCS-MRI scheme is unable to reconstruct MR images exactly at low sampling rate. However, the performance of MR images by WCS-MRI is better than that by FCS-MRI. In Fig. 11, WCS-MRI can reconstruct MR images without apparent artifacts and loss of high-resolution components, while in FCS-MRI, there are much apparent artifacts and loss of high-resolution components in reconstruction results. As expected, improved SNR leads to improved reconstruction quality for these two schemes. These figures indicate that WCS-MRI performs better than FCS-MRI regardless of noise level. 4.2.3.Quantitative evaluation of image reconstruction performanceThe fully sampled Fourier-encoded MR images in Fig. 4 are used as the gold standards to evaluate the image reconstruction performance. In order to measure the similarity between the reconstructed image and the gold-standard image , the PSNR and SSIM index are used.43,44 PSNR is the most widely used image quality assessment metric and has clear physical meaning The SSIM metric is used to measure visual reconstruction quality by capturing the similarity between the original image and the reconstructed image. SSIM models any distortion as a combination of three different factors: loss of correlation, luminance distortion, and contrast distortion. The dynamic range of SSIM is . SSIM represents the degree of similarity between and . The higher the SSIM is, the more similar and are. The maximum value of 1 is achieved only if . Figures 12 and 13 show PSNR and SSIM as functions of data sampling rate in the ideal condition, respectively. Fig. 12PSNRs of reconstruction results at different sampling rates (a) phantom, (b) brain image 1 and (c) brain image 2. 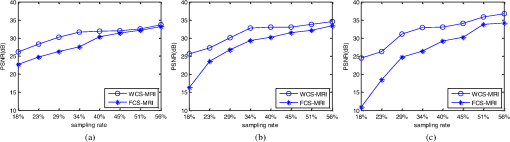 Fig. 13SSIMs of reconstruction results at different sampling rates for (a) phantom, (b) brain image 1 and (c) brain image 2. 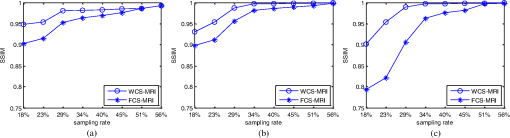 As shown in Figs. 12 and 13, with , WCS-MRI can achieve very good imaging quality with and in all tested images. In low sampling rates (i.e., ), WCS-MRI outperform FCS-MRI with higher PSNRs and SSIMs. Even with , PSNR in WCS-MRI can reach more than 25 dB and SSIM more than 0.9. With all tested sampling rates and images, PSNRs and SSIMs are higher in WCS-MRI than in FCS-MRI. Figures 14 and 15 show the median PSNR and SSIM as functions of noise levels (, 20 dB, 30 dB and 40 dB) when sampling rate is 34%. WCS-MRI scheme achieves a better reconstruction performance with higher value of PSNR and SSIM than FCS-MRI scheme at each noise level. 4.2.4.Improvement in CS reconstructionThe use of sparse-encoding in wavelet-encoded -space in this work was motivated by the desire to improve RIP constants to achieve good CS reconstruction. It is difficult to compute RIP constants for all signal construction, but we can calculate them in the FCS-MRI scheme by Eq. (12) and in WCS-MRI scheme by Eq. (13) for each tested MR image. Table 2 shows the value of RIP constant ( is determined by SR) for each test image. When the value of is smaller, the reconstruction performance is better in the corresponding image. The value of in WCS-MRI is less than that in FCS-MRI for most conditions. This implies that WCS-MRI provides better reconstruction stability and accuracy than FCS-MRI. Table 2The RIP constants for tested images in each condition.
5.DiscussionSimulated dataset have been used to assess CS reconstruction performance in wavelet domain and Fourier domain for different sampling rates, noise and sparsity levels. CS reconstruction of MR image in wavelet domain achieved better quality than in Fourier domain, especially ringing artifact reduction in the region of edges. Based on the simulation results, PSNR and SSIM indices were computed to evaluate the performance of MR image reconstruction in ideal and noisy conditions. In the case of low-SNR and low sampling rate, WCS-MRI achieved much higher values of PSNR and SSIM than FCS-MRI. In the case of high-SNR and high sampling rate, FCS-MRI and WCS-MRI both achieved similar values. That means WCS-MRI has advantages in reduction of scan time. When the sampling rate is more than 30%, WCS-MRI reconstruction achieves . The acquisition time of MR images can be reduced significantly (by almost 70%) without much distortion when the proposed WCS-MRI method is applied. 6.ConclusionsThis work describes the development of a simulator for MRI with a wavelet-encoding scheme for stable and accurate MR image reconstruction by acquiring subsampled data below the Nyquist sampling rate following the embedded hierarchical tree structure of significant wavelet coefficients within a CS framework. According to our simulation results, the proposed scheme improves the existing Fourier-encoded CS-MRI method by introducing this specific wavelet encoding instead of commonly used random encoding in the -space data acquisition. The hierarchical wavelet tree structure is applied to select sparse encodings in order to improve the precision of the CS reconstruction by satisfying the requirement of RIP. The acquired under sampled -space contains many significant samples and the reconstruction quality of such under sampled -space is guaranteed by using the input vector of sparse wavelet coefficients in a specific CS framework, which is based on the minimization of total-variation (TV) regularization signal and a least squares measurement. Based on the flexibility and adaptivity in wavelet-encoded MR data acquisition, it is feasible to implement sparse encoding of -space data with tailored spatially selective RF excitation pulses without much modification of MRI machinery. Therefore, this proposed encoding scheme may provide a practical method for shortening the patient scan time by reducing the required number of RF excitation pulses without decreasing the resolution of reconstructed MR images. In addition, this work may be applied in future to functional MRI (fMRI), where the three- and four-dimensional data would be even more compressible than the two dimensional data used in the wavelet transform domain. For example in echo planar imaging (EPI), using wavelet-shaped slice-selection excitation pulse with sparse encoding of slice data may accelerate fMRI with improved CS reconstruction. AcknowledgmentsThis work was partially funded by an internal seed grant. The authors would like to thank Prof. J. M. Fitzpatrick and Dr. D. Yoder of the EECS Department of Vanderbilt University for kindly sharing their Fourier domain MRI simulator code with us. ReferencesZ. P. LiangP. C. Lauterbur,
“Principles of magnetic resonance imaging: a signal processing perspective,”
107
–136 IEEE Press, New York
(2000). Google Scholar
G. McGibneyet al.,
“Quantitative evaluation of several partial Fourier reconstruction algorithms used in MRI,”
Magn. Reson. Med., 30
(1), 51
–59
(1993). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
C. M. TsaiD. Nishimura,
“Reduced aliasing artifacts using variable-density -space sampling trajectories,”
Magn. Reson. Med., 43
(3), 452
–458
(2000). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
A. GreiserM. von Kienlin,
“Efficient k-space sampling by density-weighted phase-encoding,”
Magn. Reson. Med., 50
(6), 1266
–1275
(2003). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
B. MadoreG. H. GloverN. J. Pelc,
“Unaliasing by Fourier-encoding the overlaps using the temporal dimension (UNFOLD), applied to cardiac imaging and fMRI,”
Magn. Reson. Med., 42
(5), 813
–828
(1999). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
L. P. PanychP. D. JakabF. A. Jolesz,
“Implementation of wavelet-encoded MR imaging,”
J. Magn. Reson. Imag., 3
(4), 649
–655
(1993). http://dx.doi.org/10.1002/(ISSN)1522-2586 1053-1807 Google Scholar
D. M. HealyJ. B. Weaver,
“Two applications of wavelet transforms in magnetic resonance imaging,”
IEEE Trans. Inform. Theor., 38
(2), 840
–860
(1992). http://dx.doi.org/10.1109/18.119740 IETTAW 0018-9448 Google Scholar
L. P. Panych,
“Theoretical comparison of Fourier and wavelet encoding in magnetic resonance imaging,”
IEEE Trans. Med. Imag., 15
(2), 141
–153
(1996). http://dx.doi.org/10.1109/42.491416 ITMID4 0278-0062 Google Scholar
D. L. Donoho,
“Compressed sensing,”
IEEE Trans. Information Theor., 52
(4), 1289
–1306
(2006). http://dx.doi.org/10.1109/TIT.2006.871582 IETTAW 0018-9448 Google Scholar
M. Lustiget al.,
“Compressed sensing MRI,”
IEEE Signal Process. Mag., 25
(2), 72
–82
(2008). http://dx.doi.org/10.1109/MSP.2007.914728 ISPRE6 1053-5888 Google Scholar
M. LustigD. L. DonohoJ. M. Pauly,
“Sparse MRI: the application of compressed sensing for rapid MR imaging,”
Magn. Reson. Med., 58
(6), 1182
–1195
(2007). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
U. GamperP. BoesigerS. Kozerke,
“Compressed sensing in dynamic MRI,”
Magn. Reson. Med., 59
(2), 365
–373
(2008). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
Z. WangG. R. Arce,
“Variable density compressed image sampling,”
IEEE Trans. Image Process., 19
(1), 264
–270
(2010). http://dx.doi.org/10.1109/TIP.2009.2032889 IIPRE4 1057-7149 Google Scholar
Y.-C. KimS. S. NarayananK. S. Nayak,
“Accelerated three-dimensional upper airway MRI using compressed sensing,”
Magn. Reson. Med., 61
(6), 1434
–1440
(2009). http://dx.doi.org/10.1002/mrm.v61:6 MRMEEN 0740-3194 Google Scholar
C. O. Schirraet al.,
“Toward true 3D visualization of active catheters using compressed sensing,”
Magn. Reson. Med., 62
(2), 341
–347
(2009). http://dx.doi.org/10.1002/mrm.v62:2 MRMEEN 0740-3194 Google Scholar
D. Lianget al.,
“Accelerating SENSE using compressed sensing,”
Magn. Reson. Med., 62
(6), 1574
–1584
(2009). http://dx.doi.org/10.1002/mrm.v62:6 MRMEEN 0740-3194 Google Scholar
M. Seegeret al.,
“Optimization of k-space trajectories for compressed sensing by Bayesian experimental design,”
Magn. Reson. Med., 63
(1), 116
–126
(2010). MRMEEN 0740-3194 Google Scholar
L. JacquesP. Vandergheynst,
“Compressed sensing: when sparsity meets sampling,”
Optical and Digital Image Processing–Fundamentals and Applications, 507
–527 Wiley-VCH, Hoboken, NJ
(2011). Google Scholar
J. P. HaldarD. HernandoZ. P. Liang,
“Compressed sensing in MRI with random encoding,”
IEEE Trans. Med. Imaging, 30
(4), 893
–903
(2011). http://dx.doi.org/10.1109/TMI.2010.2085084 ITMID4 0278-0062 Google Scholar
D. Lianget al.,
“Toeplitz random encoding MR imaging using compressed sensing,”
in Proc. IEEE Int. Symp. Biomed. Imaging,
270
–273
(2009). Google Scholar
H. WangD. LiangL. Ying,
“Pseudo 2D random sampling for compressed sensing MRI,”
in Proc. IEEE Engineering in Medicine and Biology Conference,
2672
–2675
(2009). Google Scholar
S. DeutschA. AverbuchS. Dekel,
“Adaptive compressed image sensing based on wavelet modeling and direct sampling,”
in Proc. 8th International Conference on Sampling Theory and Applications,
(2009). Google Scholar
A. SoniJ. Haupt,
“Efficient adaptive compressive sensing using sparse hierarchical learned dictionaries,”
in Proc. Asilomar Conf. on Signals, Systems, and Computers,
1250
–1254
(2011). Google Scholar
J. HauptR. CastroR. Nowak,
“Distilled sensing: adaptive sampling for sparse detection and estimation,”
IEEE Trans. Inform. Theor., 57
(9), 6222
–6235
(2011). http://dx.doi.org/10.1109/TIT.2011.2162269 IETTAW 0018-9448 Google Scholar
E. Arias-CastroE. J. CandesM. Davenport,
“On the fundamental limits of adaptive sensing,”
(2011). Google Scholar
J. S. WalkerT. Q. Nguyen,
“Wavelet-based image compression,”
The Transform and Data Compression Handbook, 267
–312 CRC Press, Boca Raton
(2000). Google Scholar
D.S. TaubmanM. W. Marcellin,
“JPEG 2000: image compression fundamentals, standards and practice,”
Kluwer International Series in Engineering and Computer Science, Kluwer Academic, Norwell, MA
(2002). Google Scholar
J. M. Shapiro,
“Embedded image coding using zerotrees of wavelet coefficients,”
IEEE Trans. Signal Process., 41
(12), 3445
–3459
(1993). http://dx.doi.org/10.1109/78.258085 ITPRED 1053-587X Google Scholar
C. LaM. N. Do,
“Signal reconstruction using sparse tree representation,”
Proc. SPIE, 5914 273
–283
(2005). Google Scholar
D. A. Yoderet al.,
“MRI simulator with object-specific field map calculations,”
Magnetic Reson. Imaging, 22
(3), 315
–328
(2004). http://dx.doi.org/10.1016/j.mri.2003.10.001 MRIMDQ 0730-725X Google Scholar
H. Benoit-Cattinet al.,
“The SIMRI project: a versatile and interactive MRI simulator,”
J. Magn. Reson., 173
(1), 97
–115
(2005). http://dx.doi.org/10.1016/j.jmr.2004.09.027 JOMRA4 0022-2364 Google Scholar
S. Saekhoet al.,
“Small tip angle three dimensional tailored radiofrequency slab-select pulse for reduced B1 inhomogeneity at 3T,”
Magn. Reson. Med., 53
(2), 479
–484
(2005). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
H. GudbjartssonS. Patz,
“The Rician distribution of noisy MRI data,”
Magn. Reson. Med., 34
(6), 910
–914
(1995). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
J. Sijberset al.,
“Estimation of the noise in magnitude MR images,”
Magn. Reson. Imag., 16
(1), 87
–90
(1998). MRMEEN 0740-3194 Google Scholar
R. Chartrand,
“Exact reconstruction of sparse signals via nonconvex minimization,”
Sig. Proc. Lett., 14
(10), 707
–710
(2007). http://dx.doi.org/10.1109/LSP.2007.898300 IESPEJ 1070-9908 Google Scholar
W. DaiO. Milenkovic,
“Subspace pursuit for compressive sensing signal reconstruction,”
IEEE Trans. Inf. Th., 55
(5), 2230
–2249
(2009). http://dx.doi.org/10.1109/TIT.2009.2016006 IETTAW 0018-9448 Google Scholar
Y. TsaigD. L. Donoho,
“Extensions of compressed sensing,”
Signal Process., 86
(3), 533
–548
(2006). http://dx.doi.org/10.1016/j.sigpro.2005.05.028 SPRODR 0165-1684 Google Scholar
E. J. Candes,
“The restricted isometry property and its implications for compressed sensing,”
C. R. l’Academie des Sci., 346
(9), 589
–592
(2008). http://dx.doi.org/10.1016/j.crma.2008.03.014 1631-073X Google Scholar
P. KoiranA. Zouzias,
“On the certification of the restricted isometry property,”
(2011). Google Scholar
E. J. CandesM. B. Wakin,
“An introduction to compressive sampling,”
IEEE Signal Process. Mag., 25
(2), 21
–30
(2008). http://dx.doi.org/10.1109/MSP.2007.914731 ISPRE6 1053-5888 Google Scholar
H. GachC. TanaseF. Boada,
“2D & 3D Shepp-Logan phantom standards for MRI,”
in Proc. 19th Int. Conf. Syst. Eng.,
521
–526
(2008). Google Scholar
D. L. CollinsA. P. ZijdenbosV. Kollokian,
“Design and construction of a realistic digital brain phantom,”
IEEE Trans. Medical Imaging, 17
(3), 463
–468
(1998). http://dx.doi.org/10.1109/42.712135 ITMID4 0278-0062 Google Scholar
Z. WangA. C. Bovik,
“A universal image quality index,”
IEEE Signal Process. Lett., 9
(3), 81
–84
(2002). http://dx.doi.org/10.1109/97.995823 IESPEJ 1070-9908 Google Scholar
Z. Wanget al.,
“Image quality assessment: from error visibility to structural similarity,”
IEEE Trans. Image Processing, 13
(4), 600
–612
(2004). http://dx.doi.org/10.1109/TIP.2003.819861 IIPRE4 1057-7149 Google Scholar
Biography Zheng Liu received the BSEE degree from Northwestern Polytechnical University, Xi’an, Shannxi, China, in 2003 and MSEE and PhD degrees from Tianjin University, Tianjin, China, in 2006 and 2009, respectively. He was a postdoctoral fellow in the Department of Electrical and Computer Engineering, Texas Tech University (Lubbock, Texas) (May 2009 to November 2012), where his work focused on MRI simulation, pulse sequence programming, MRI/fMRI signal/image processing and quality testing. Currently, he is working as a postdoctoral researcher II in the Department of Radiology & Advanced Imaging Research Center (AIRC), UT Southwestern Medical Center at Dallas, (Dallas, Texas), where his work focused on development of novel imaging protocols, CEST-based contrast methods for diagnosis, especially in CEST to image important parameters (e.g., pH, metabolite level) for early detection and monitoring of diseases, such as osteoarthritis and disc degeneration. His interests include optical imaging systems, magnetic resonance imaging (MRI), image processing, and real-time embedded systems.  Brian Nutter received BSEE and PhD degrees from Texas Tech University, Lubbock, Texas, in 1987 and 1990, respectively. He is an associate professor and associate chairman in the Department of Electrical and Computer Engineering, Texas Tech University (Lubbock, Texas). He was a software/electronics designer and manager in two rapid prototyping companies, 3-D Systems (1990 to 1992) and Soligen (1992 to 1998), where his work provided key technologies in data representation, motion control, and system design. He was vice president of engineering at WillowBrook Technologies, a manufacturer of digital telephone systems, from 1998 to 2002, where he provided the technical direction for a very innovative telephony solution. He was a member of the startup team for both WillowBrook and Soligen. His interests include telecommunications, networks, biomedical signal and image processing, rapid prototyping, and real-time embedded systems.  Sunanda Mitra received the BSc degree in 1955 and the MSc degree in 1957, both in physics, from Calcutta University, Calcutta, India, and the DSc (Doctors der Naturwissenshaften) degree in physics from Marburg University, Marburg, Germany, in 1966. She has served as the director of Computer Vision and Image Analysis Laboratory, Department of Electrical and Computer Engineering, Texas Tech University (TTU), Lubbock, Texas, since 1988. Prior to joining TTU in 1984, she worked as a research scientist at TTU’s School of Medicine (1969 to 1983) and as a visiting faculty member (1983 to 1984) at the Mount Sinai School of Medicine, New York. She also held a faculty position (1960 to 1964) at Lady Brabourne College, Calcutta, India. She served on the Board of Scientific Counselors of the National Library of Medicine at the National Institutes of Health (USA) from 1997 to 2000. She holds the P.W. Horn Professorship at TTU and has published over 160 scientific articles, including archival journal papers, invited papers, and book chapters. Her specialization includes medical image segmentation and analysis, data compression, 3-D modeling from stereo vision and pattern recognition. She has chaired the Technical Committee of Computational Medicine of the IEEE Computer Society. She is also on the Program Committee of the International Medical Imaging Symposium on “Image Processing” sponsored by SPIE. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||